camfr_guided_modes
- ipkiss3.all.device_sim.camfr_guided_modes(material_stack, wavelengths, pml_thickness=0.0)
Calculates the neff for guided modes for a material stack using the camfr mode solver.
- Parameters:
- material_stack: MaterialStack object
The stack of ipkiss materials to calculate the modes for.
- wavelengths: array_like
An array of wavelengths to calculate the modes for.
- pml_thickness: float
Thickness of the complex Perfectly Matched Layer to use.
- Returns:
- guided_modes: dict
Returns a dictionary that maps the mode names onto a numpy array that contains the wavelength and the corresponding mode effective index.
Examples
import si_fab.all as pdk import ipkiss3.all as i3 import numpy as np import matplotlib.pyplot as plt from pysics.basics.material.material_stack import MaterialStack # we define a MaterialStack mstack = MaterialStack( name="150nm Si", materials_heights=[ (i3.TECH.MATERIALS.SILICON_OXIDE, 0.500), (i3.TECH.MATERIALS.SILICON, 0.150), (i3.TECH.MATERIALS.AIR, 1.070) ] ) # use the MaterialStack method to plot it mstack.visualize()
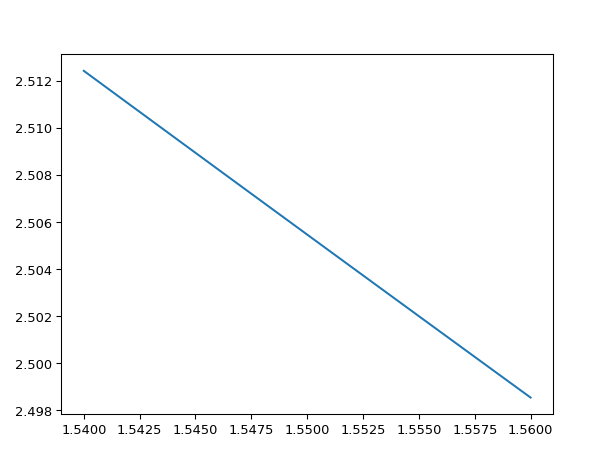
wavelengths = np.linspace(1.54, 1.56, 10) neffs = i3.device_sim.camfr_guided_modes(mstack, wavelengths) # Now we plot the zeroth-order mode: TE0 = neffs['TE0'] # TEO is 'structured numpy array', this means it has fields # in this case it has a wavelength and n_eff field. wls = TE0['wavelength'] # n_eff is a complex number neffs = TE0['n_eff'].real plt.plot(wls, neffs) plt.show()
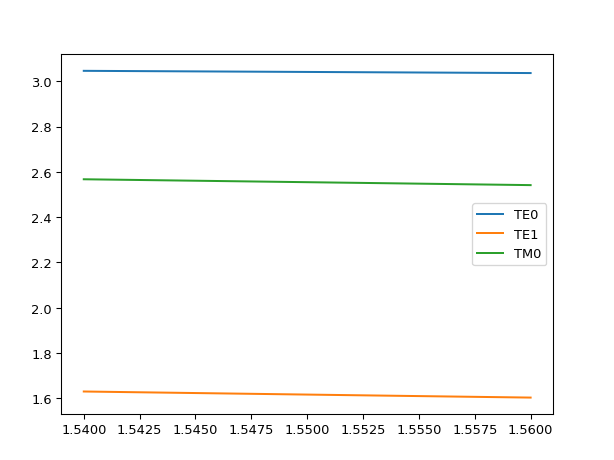
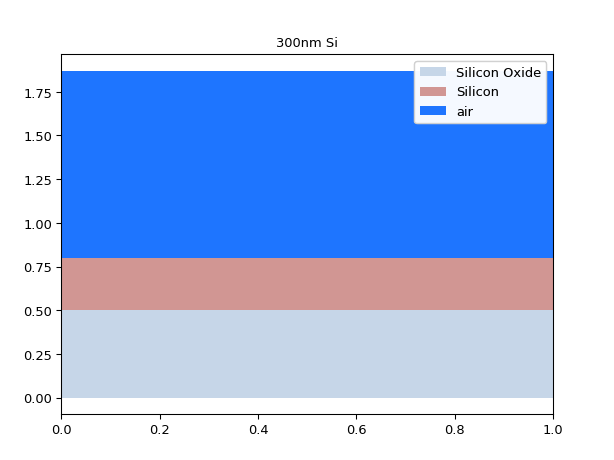
# If we make the core higher # we should see higher order modes appear mstack = MaterialStack( name="300nm Si", materials_heights=[ (i3.TECH.MATERIALS.SILICON_OXIDE, 0.500), (i3.TECH.MATERIALS.SILICON, 0.300), (i3.TECH.MATERIALS.AIR, 1.070) ] ) mstack.visualize()
results = i3.device_sim.camfr_guided_modes(mstack, wavelengths) for mode_name, neffs in results.items(): plt.plot(neffs['wavelength'], neffs['n_eff'].real, label=mode_name) plt.legend(results.keys()) plt.show()
Sometimes you’ll want to write/load the data to/from a file. To allow reading it from a different application or just to avoid running the simulation multiple times.
# With Python's builtin pickle module it's easy to write # and load the data to/from a data file. import si_fab.all as pdk from pysics.basics.material.material_stack import MaterialStack import ipkiss3.all as i3 import numpy as np mstack = MaterialStack( name="300nm Si", materials_heights=[ (i3.TECH.MATERIALS.SILICON_OXIDE, 0.500), (i3.TECH.MATERIALS.SILICON, 0.300), (i3.TECH.MATERIALS.AIR, 1.070) ] ) wavelengths = np.linspace(1.54, 1.56, 10) results = i3.device_sim.camfr_guided_modes(mstack, wavelengths) # to write to the file import pickle with open('out.dat', 'wb') as out: pickle.dump(results, out) fin = open('out.dat', 'rb') results_new = pickle.load(fin) fin.close() # in case you want to export to a textual format, e.g a csv file # you can use np.savetxt, though this requires a bit more work: def savetocsv(filename, results): results_items = results.items() data = np.hstack([val for _, val in results_items]) dtype = [ ('mode_name', 'S10'), ('wavelength', np.float64), ('neff_real', np.float64), ('neff_imag', np.float64) ] results_np = np.zeros( len(data), dtype=dtype ) offset = 0 for mode_name, result in results_items: results_np['mode_name'][offset:offset + len(result)] = mode_name offset += len(result) results_np['wavelength'] = data['wavelength'] results_np['neff_real'] = data['n_eff'].real results_np['neff_imag'] = data['n_eff'].imag np.savetxt(filename, results_np, delimiter=',', fmt=['%s', '%.18e', '%.18e', '%.18e']) savetocsv('guided_modes.csv', results) # to load the csv data again, you can use np.loadtxt: def loadfromcsv(filename): # we assume the same order as used in the savetocsv dtype = [ ('mode_name', 'S10'), ('wavelength', np.float64), ('neff_real', np.float64), ('neff_imag', np.float64) ] raw_data = np.loadtxt(filename, dtype=dtype, delimiter=',') mode_names = np.unique(raw_data['mode_name']).tolist() results = {} for mode_name in mode_names: mask = raw_data['mode_name'] == mode_name mode_data = raw_data[mask][['wavelength', 'neff_real', 'neff_imag']] results[mode_name] = np.rec.fromarrays( [mode_data['wavelength'], mode_data['neff_real'] + 1j * mode_data['neff_imag']], names='wavelength, n_eff' ) return results loaded_results = loadfromcsv('guided_modes.csv') # Note that loadtxt filters out lines starting with '#'. # You can use this to add comments to your data: with open('guided_modes_comments.csv', 'w') as csv_out: csv_out.writelines([ '# This file contains n_eff data for our material stack\n', '# mode_name, wavelength, neff_real, neff_imag\n' ]) savetocsv(csv_out, results) # comments will be filtered out: loaded_results = loadfromcsv('guided_modes_comments.csv')
A MaterialStack might require preprocessing before calculating the modes. The code snippet below illustrates how you can define a function that clip a material stack between a z_min and z_max value.
import si_fab.all as pdk from pysics.basics.material.material_stack import MaterialStack import ipkiss3.all as i3 import numpy as np mstack = MaterialStack( name="300nm Si", materials_heights=[ (i3.TECH.MATERIALS.SILICON_OXIDE, 0.500), (i3.TECH.MATERIALS.SILICON, 0.300), (i3.TECH.MATERIALS.AIR, 1.070) ] ) mstack.visualize()
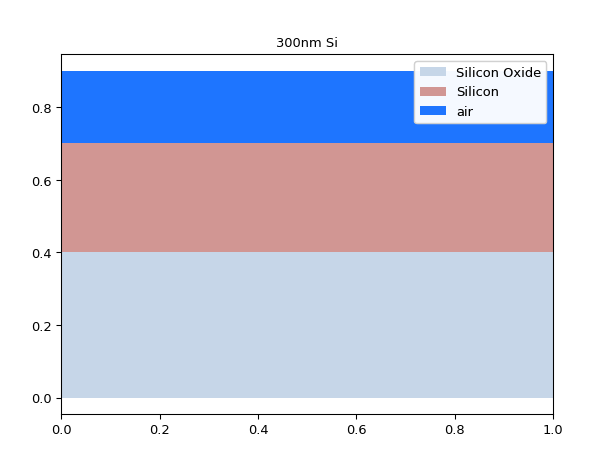
mstack_b = mstack.clip_copy(z_min=0.1, z_max=1.0) mstack_b.visualize()
Using negative values for z_min will make the material stack grow. If you use a z_max value higher than the top of the material stack, it will grow as well.
mstack_c = mstack_b.clip_copy(z_min=-1, z_max=2.0) mstack_c.visualize()